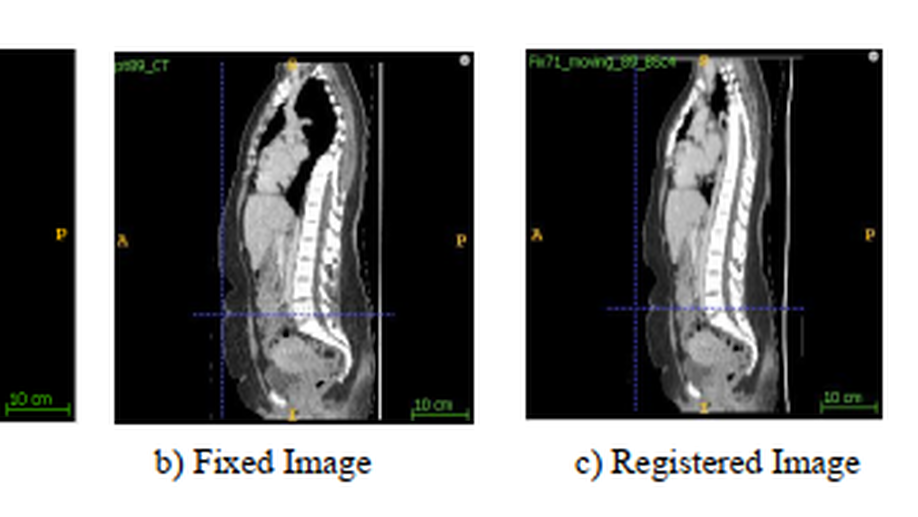
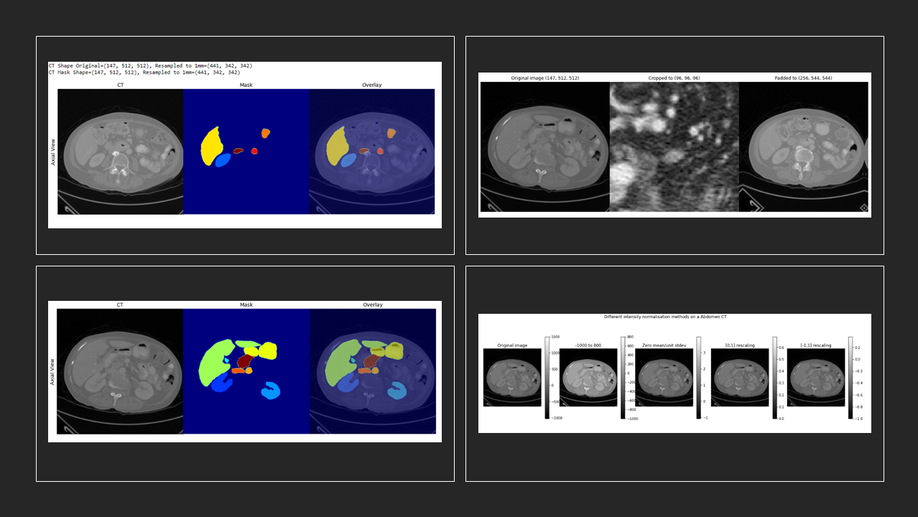
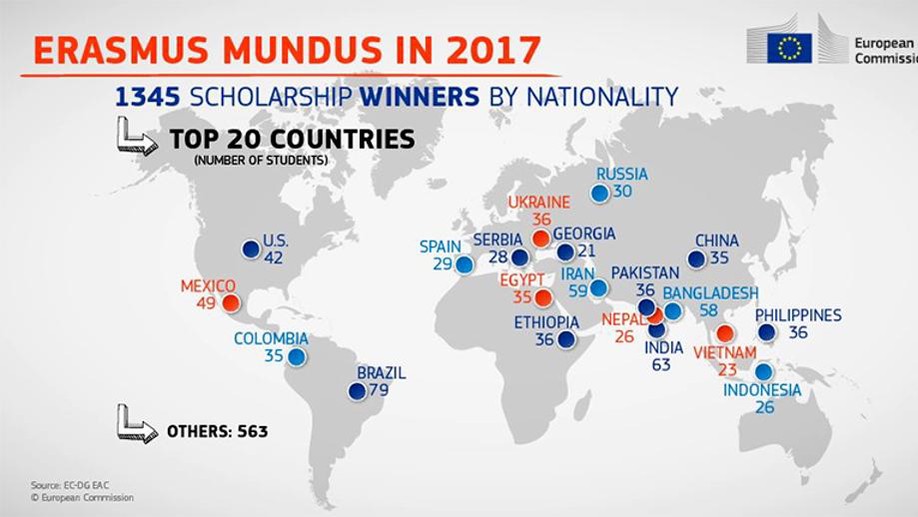
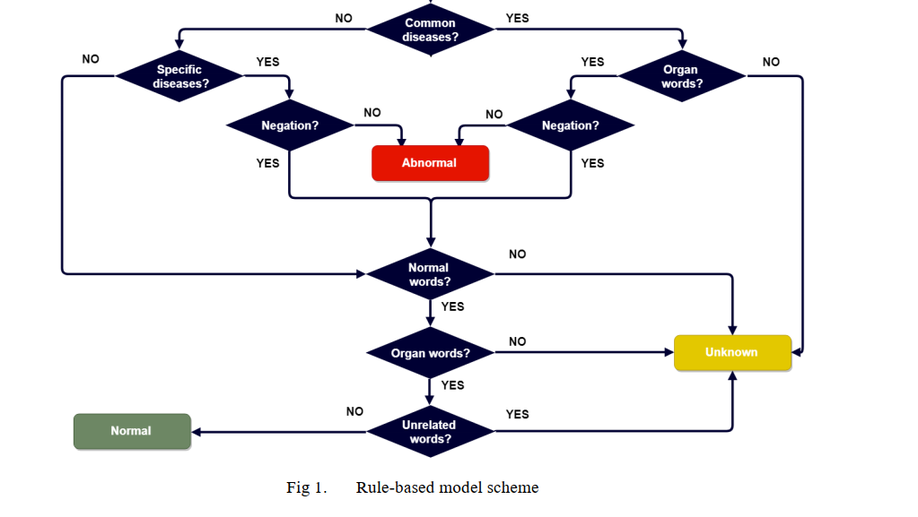
Fakrul Islam Tushar
Associate in Research
Duke University Medical Center
Erasmus Mundus Joint Master Degree in Medical Imaging and Applications
Biography
Fakrul Islam tushar is a Medical Imaging & Computer Vision Engineer, primarily engaged in research, computer-aided diagnosis and healthcare innovation using machine learning, neural networks, and image analysis driven solutions. He graduated from the Erasmus+ Joint Master in Medical Imaging and Applications and is currently a Post-Graduate Research Associate at the Carl E. Ravin Advanced Imaging Laboratories (RAI Labs), Duke University Medical Center, USA. He earned a Bachelor of Science degree in Electrical and Electronics Engineering at American International University Bangladesh (AIUB). He is a recipient of the “Cum Laude” distinction and academic honor at the 17th Convocation Ceremony of AIUB, “Dean’s Award” for his final-year undergraduate research project, the European Union: Erasmus+ Grant and the Duke University: Masters Thesis Grant. His past affiliations include IEEE AIUB Student Branch, Teach For Bangladesh and “Literacy Through Leadership”.
Interests
- Artificial Intelligence
- Medical Image Processing and Analysis
- Image Segmentation
- Deeplearning
- NLP
Education
Associate in Research, 2019-present
Duke University Medical Center
Masters in Medical Imaging and Applications, 2017-2019
Erasmus Mundus Joint Master ( University of Burgundy; University of Cassino; University of Girona, Duke University)
BSc in Electrical & Electronic Engineering, 2013-2017
American International University Bangladesh